This article was featured in Library Notes, #47 (Spring 2008).
Well, spring is here and that means that new things are springing up all over. This includes new resources from the NCBI (National Center for Biotechnology Information). Recently NCBI introduced their new Sequence Viewer, which is replacing the old graphical view of a sequence accessed from the Entrez Nucleotide or Entrez Protein databases, though my favorite access point is from the Entrez Gene record for the gene I am viewing at any given time.
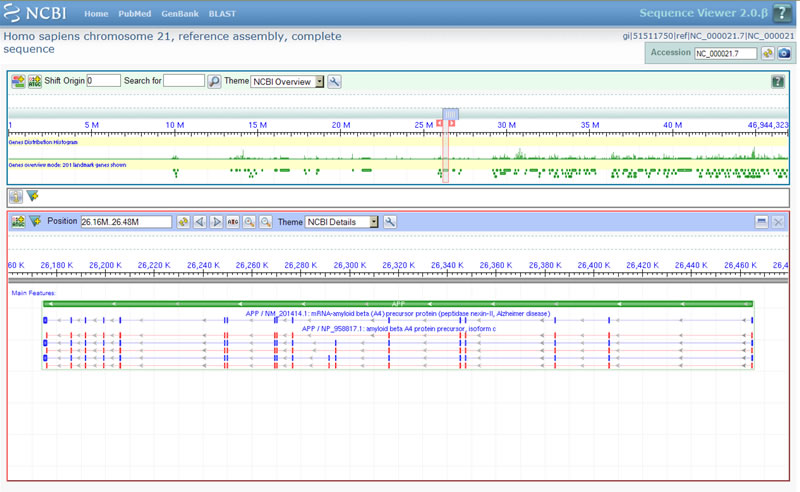
Practically everything in the new viewer is interactive: you can get a wide or narrow horizontal view of a chromosome with genes visible. You can view genes' isoforms, sequences and SNP profiles. Take a look at the intro pages for Sequence Viewer.
NCBI has also released a new version of the Conserved Domains Database (CDD) with many more interactive and informative features than ever before. When you run a BLAST, the new CDD will now show you the fold family and superfamily (if available) for any conserved domains found in your query.
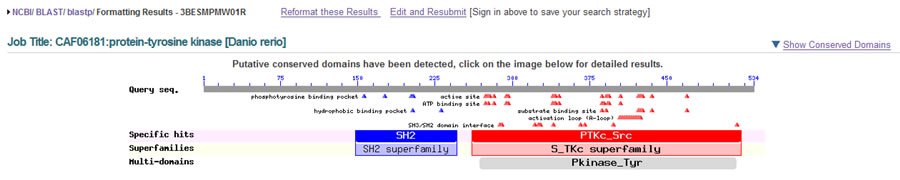
You will see sites of conserved features such as activation loops, binding sites for ATP, ligands and other substrates. You can choose to view the structures that are available for those domain family members, and see a phylogenetic tree for the family.
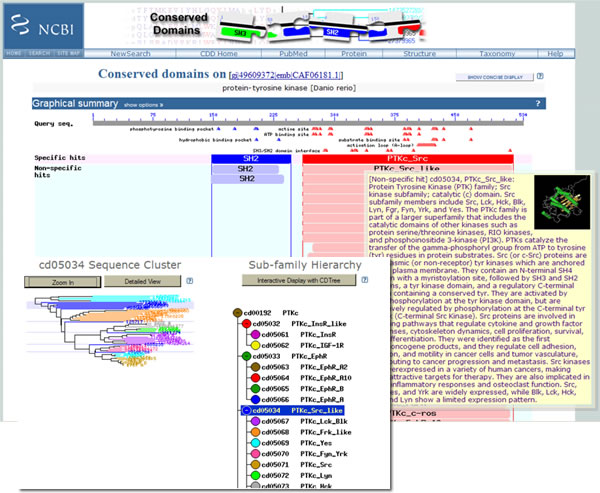
For an overview of all the features of the new CDD, see the help pages.
Finally, I'd like to get some input from the research community. I am looking at a number of pathway analysis software suites, such as Ingenuity Pathways Analysis and GeneGo Metacore. Many of these packages have features that allow users to integrate their own high-throughput data and to customize the pathway analysis. Take a look at some of these companies' products:
- Ingenuity Pathways Analysis http://www.ingenuity.com/
- GeneGo MetaCore http://www.genego.com/metacore.php
Please e-mail me or leave a comment to this blog entry and let me know if you need such software available for your research. With NU's new clinical and translational center and funding I suspect there will be increased demand for such packages, and I'd like to get some idea of what our need is here at Northwestern. Just send a quick note to me indicating if you would use pathway analysis tools and what package you would prefer, if any. You do not need to limit your preference to the two I listed above. They are there as examples for you to investigate.
Don't forget, I am available to provide instruction and assistance in the use of a variety of literature and molecular databases and tools. Please feel free to drop me a message any time you need to.
Pamela Shaw
Biosciences Librarian
312-503-8689
Updated: September 25, 2023