This article was featured in Library Notes #48 (Fall 2008).
A common question asked of bioinformatics consultants in libraries is "What can I use for SNP annotation?" The discouraging answer is that there are very few tools that satisfy most users' demands for features in SNP annotation. There are a few, though, that come close. One of these is Snap.

Snap stands for "SNP Annotation Platform", and is hosted by the Institute for Human Genetics at Aarhus University of Denmark. While there are other online and desktop SNP annotation tools, this one is good because it is updated regularly and draws its information from Ensembl, KEGG, UniProt, UCSC, OMIM and Pfam databases.
Genes can be searched in Snap by gene name, marker, clone or SNP ID. Search results are returned with known features and characteristics listed for the gene under investigation, complete with links to the appropriate database from which the evidence was taken.
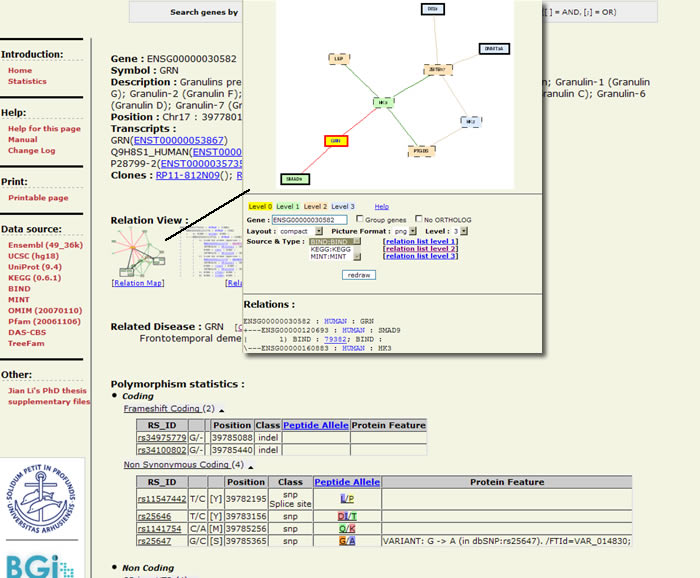
Features of the search result screen include a "relation view" that opens in a new window, illustrating the network of interactions between your gene of interest and other genes. The graphic display also includes all known polymorphism classes, the residues for which the SNPs code and any resultant protein variants.
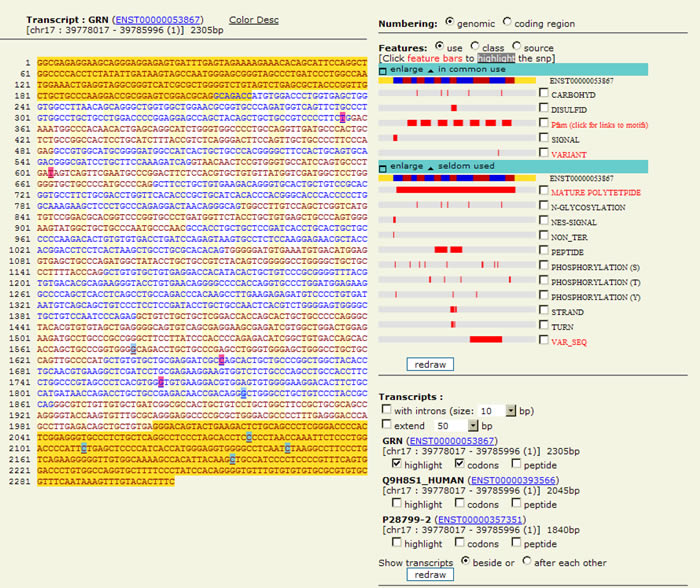
Snap includes a now-familiar graphic style of gene representation, with the sequence color-coded by region (5' and 3' UTRs, coding region, etc.), but this platform allows users to redraw the sequence graphic by changing features of the introns, phosphorylation and N-glycosylation sites and other features in the gene.
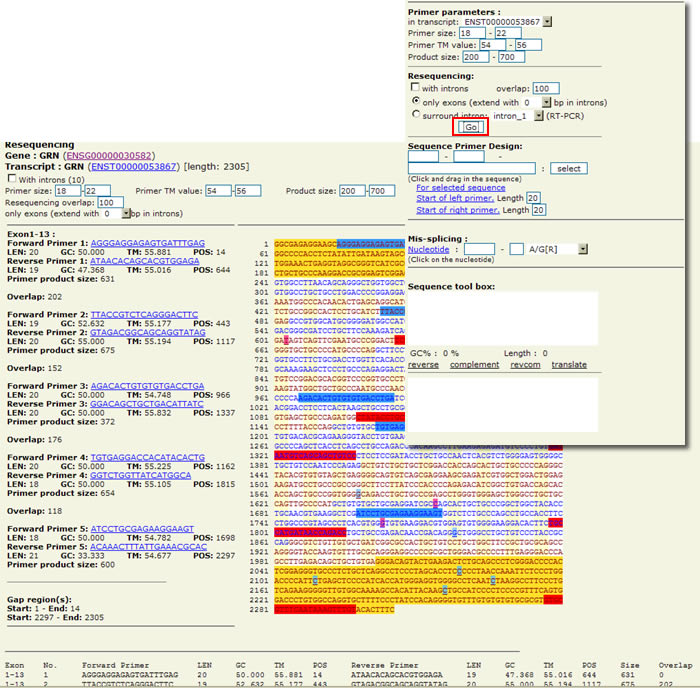
Finally, Snap allows users to view forward or reverse primer sequences for the most significant regions of the gene, or to select a portion of the sequence, by clicking and dragging, for which Snap will recommend primers in a separate browser window.
This flexibility, combined with links to sources of evidence in OMIM, KEGG and Pfam, provides one of the most complete free-access SNP annotation platforms available.
Try it out!
Do you have suggestions for other sites you'd like to see featured in the Biosciences Blog? Send a recommendation or a comment to me:
Pamela Shaw
Biosciences Librarian
312-503-8689
Updated: September 25, 2023